Hi,
Thanks as always for the insightful questions.
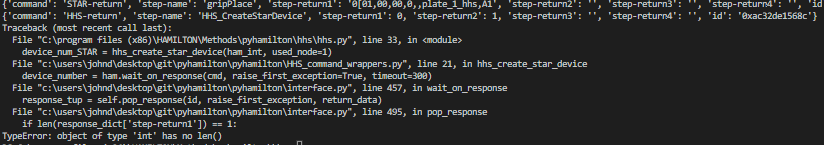
Edit: I seem to have managed to get something working. Not entirely sure what I did, because I think all I did was update my fork to match the official pyhamilton repo. Regardless, my questions above are still mostly valid as I’m still not certain as to what the return_fields represent!
Funny enough I actually just fixed this, which is why it started to work for you. This is an excellent demonstration of good open-source practices!
To your question: Some of our wrappers include a field return_data such that HamiltonInterface.wait_on_response(..., return_data=['step-return2']). This means that instead of performing normal error checking (or ignoring) a field in the JSON response string from the Venus method, we actually return the data from the wrapper function. This is used if we have a sensor integrated into PyHamilton that is called from Venus.
If you don’t return sensor data or something similar that you want to process in Python, there’s no need to have return_data be anything but the default value which is None.
Another question I have is regarding the STAR_OEM_Test.lay file. It always incurs indirect changes when I test changes to the source code. Should I include STAR_OEM_Test.lay with my commits, or should I omit it?
That’s a great point that has annoyed me for some time as well. I think we can add the .lay to a .gitignore file and avoid shipping it with the core library. It’s only been a headache for users.
One final question on the general topic of the STAR_OEM files… Why are there so many of them and what is their purpose? We have STAR_OEM, STAR_OEM_Toolkit, STAR_OEM_Test, STAR_OEM_NoFan, STAR_OEM_wFan  I’m interested to know how they all tie together!
I’m interested to know how they all tie together!
So, STAR_OEM_noFan.hsl is of course the PyHamilton universal method. ...Toolkit is a sub-method library that we use to hold wrapper functions that we import into the universal method. ...wFan.hsl is a slightly out of date universal method that contains functions for interacting with a HEPA fan.
My lab (Sculpting Evolution, which invented PyHamilton), uses the HEPA fans for our methods, but you can’t even open a method containing a fan function if your machine/ software is not configured with a fan. I don’t know how to change this, and no other piece of equipment is like this. So we have two universal methods. noFan is the default that almost every PyHamilton user in the world uses, and wFan is I think only used by Sculpting Evolution.
There’s a bunch of other methods in VENUS_Method that are not part of this explanation. I think some of those could be removed without consequence, but I don’t actually know, and I haven’t felt compelled to try.