Hey all,
I exported a method (method 1) developed on star A, imported it on star B (did not replace the existing library files and so on). The Venus’ versions on the robots are different, but everything worked fine (method 1) in both simulation and real run (transferring serum and methanol).
Star A works fine (all methods) and has no problems when importing methods from different different Venus versions whatsoever.
However, star B has now started to throw a specific error when running on our main routine method (NOT method 1): TADM bands out of tolerance - even though TADM is turned off for this particular step.
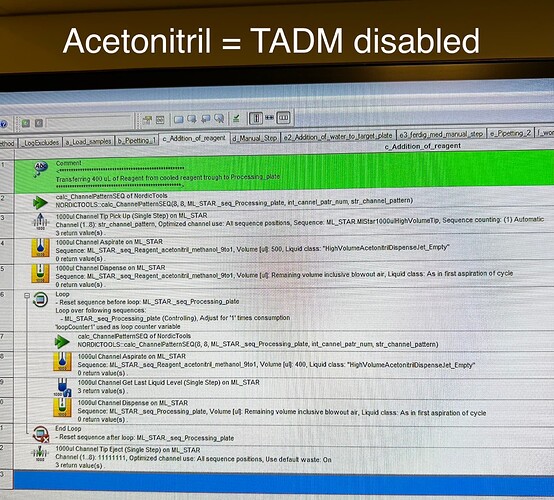
Concrete step: wet tips with 500uL of 9:1 acetonitrile:methanol, then aspirate 400 uL of the same solution, jet dispense and repeat for whole sequence. The error occurs when wetting/aspirating.
Note: high humidity (60%), but the method works on star A which is exposed to the same humidity levels.
Any ideas?
Thanks in advance!
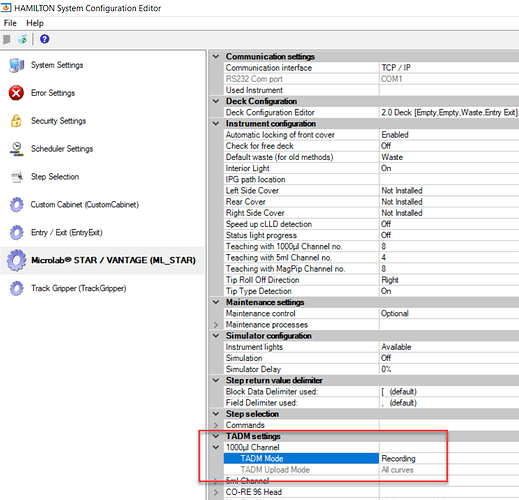
It sounds like you have TADM set to recording mode on one install, and monitoring mode on the other. That error can only be triggered when using a TADM enabled liquid class with TADM mode set to monitoring (which is a global setting defined in system configuration editor).
This error occurs when the live pressure value reported by the pipette heads internal pressure sensor exceeds or undershoots defined upper and lower pressure guard bands for the aspiration or dispense. This also occurs if guard bands are not defined in the liquid class, at the volume being used for aspiration and/or dispense. Again, these errors only occur when TADM mode is set to ‘monitoring’.
More detail regarding this topic is discussed in the following post:
https://forums.pylabrobot.org/t/detecting-a-clog/1237/2?u=nickhealy_hamilton
Is the intent to have TADM set to monitoring? If this was set by mistake, simply change to ‘recording’ mode.
Thanks.
-Nick
2 Likes
Hey Nick, thanks again for the quick reply!
The intent is to have TADM set to monitoring.
But not for this liquid (acetonitrile) (as far as I can see in the method delivered by Hamilton). What I mean is that for acetonitrile we (aka the delivered method by Hamilton) do not have recorded any bands and did not enable this feature when using the standard liquid class (acetonitrile) in our method.
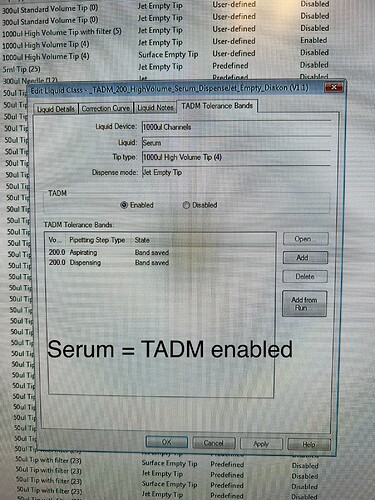
Again: this error does not occur with serum (TADM on) but with acetonitrile (TADM off).
Is it now right to assume that the error might occur due to physical parameters (“bad”/too different acetonitrile/MeOH solution, humidity) and because I imported a method from another robot (and might have messed up some library hidden some where deep)?
The method has been working fine for a long time, even after I imported methods from the other robot. But two days in a row might indicate a more serious problem which should be solved.
If you think I should reach out to Hamilton Nordics for help, let me know! I will first run som tests and see.
I hope it has something to do with the solution and not with the system. I will conduct some more tests tomorrow and let you and the community know if the error disappears by itself :):):)
@NickHealy_Hamilton
First I was working on another, new method in the method editor. I work in a hospital, plus I don’t want to mess up the delivered Hamilton method, so I decided to save this new method in a “safe” and extern (not C) directory (not sure about all the specifics and setup of this directory).
Then I was about to test the Hamilton routine method, but suddenly I received an SQL database error.
This has not happened to me before and I could not see how I provoked this error, so I restarted the pc and robot.
After that I tested the delivered Hamilton routine method and everything worked fine, despite same humidity and temperature levels as the two previous days! (star b)
So I was wondering if one should always create/edit new methods in the local C drive in order to be safe?
I strongly recommend keeping all method dependent Hamilton files (method, libraries, labware, custom dependencies) within the HAMILTON folder. Once dependent files start getting saved to non-standard drive locations (especially networked drives only your organization has access to), things can get quite messy in terms of organization, external support and export/import to other PCs/systems.
Additionally, if all installation and method-related files are within the same folder, it is much easier to backup everything needed to run a production system, or restore from backup in the case of a reinstallation or software upgrade. If files are all over the place, it can be especially hard to ensure everything was restored.
You can only impact the functionality of another method by modifying one of its dependent files (or the method itself). In most cases, this would be a custom library (SMT), labware definition, or shared deck layout file (if applicable). If you need to make modifications to a file that has not been localized to a single method and may be shared, then make a new copy and use that file for development.
In the case that something does get overwritten, then you can always restore from a backup PKG.
The SQL server error encountered when starting the run is not related to the generation of new Hamilton files outside of the C drive. If it happens again, more detail regarding the context of the error would be needed as there are numerous types of SQL server connection errors that have different causes.
Thanks.
-Nick
2 Likes
Thanks a lot!
I will do as you say.
@NickHealy_Hamilton
Some more info:
After being in contact with our local Hamilton folks we have found out that one of the runs did not execute properly (I probably aborted the run after seeing some error or closed the run control before receiving an “End method.”).
This lead to the anti-droplet-control (which I used in the new method which I was developing) not turning off properly.
This again then probably affected the TADM settings in our main routine method and resulted in an error.
Restarting the pc (and robot) leads to everything working fine again.
1 Like
Your follow-up is greatly appreciated! Thank you for the further clarification. Glad to hear everything is good now!
2 Likes